COVID-19 Cases by Ethnicity
Tommi Suvitaival, tommi.raimo.leo.suvitaival@regionh.dk, Steno Diabetes Center Copenhagen 2020-05-19
Introduction
- County-level COVID-19 data from New York Times
- County-level population data from US Census
Load Data
Census
# Source: https://www2.census.gov/library/publications/2011/compendia/usa-counties/excel/POP01.xls",
N.sheets <- 10
data.census.loaded <-
lapply(
X = 1:N.sheets,
FUN =
function( x ) {
readxl::read_excel(
path = "H:/projects/covid19/data/US_Census/POP01.xls",
sheet = x
)
}
)
# Source: https://www2.census.gov/library/publications/2011/compendia/usa-counties/excel/Mastdata.xls
mastdata.census <-
readxl::read_excel(
path = "H:/projects/covid19/data/US_Census/Mastdata.xls",
)
data.census <- data.census.loaded[[ 1 ]]
for ( i in 2:length( data.census.loaded ) ) {
data.census <-
merge(
x = data.census,
y = data.census.loaded[[ i ]][ , -1 ],
by = "STCOU",
all = TRUE
)
}
tmp <-
match(
table = mastdata.census$"Item_Id",
x = colnames( data.census )
)
data.census.orig <- data.census
colnames( data.census )[ !is.na( tmp ) ] <-
unlist( mastdata.census[ tmp[ !is.na( tmp ) ], "Item_Description" ] )
COVID-19
data.covid.loaded <-
readr::read_csv(
file = "https://raw.githubusercontent.com/nytimes/covid-19-data/master/us-counties.csv"
)
## Parsed with column specification:
## cols(
## date = col_date(format = ""),
## county = col_character(),
## state = col_character(),
## fips = col_character(),
## cases = col_double(),
## deaths = col_double()
## )
# View( data.covid.loaded )
data.covid <-
by(
data = data.covid.loaded,
INDICES = data.covid.loaded$"fips",
FUN =
function( x ) {
as.data.frame( x[ which.max( x$"date" ), ] )
}
)
data.covid <- dplyr::bind_rows( data.covid )
data.covid <-
merge(
x = data.covid,
y = USAboundaries::state_codes,
by.x = "state",
by.y = "state_name",
all.x = TRUE,
all.y = FALSE
)
data.covid$"County_State" <-
paste(
data.covid$"county",
data.covid$"state_abbr",
sep = ", "
)
Merge
data <-
merge(
x = data.covid,
y = data.census,
by.x = "County_State",
by.y = "Area_name",
all = FALSE
)
data$"Deaths_per_Capita" <-
data$"deaths" / data$"Resident population (April 1 - complete count) 2010"
data$"Deaths_per_100k" <- data$"Deaths_per_Capita" * 100000
data$"Cases_per_Capita" <-
data$"cases" / data$"Resident population (April 1 - complete count) 2010"
data$"Cases_per_100k" <- data$"Cases_per_Capita" * 100000
Compute Share of Population
names.tested <-
colnames( data.census )[ nchar( colnames( data.census ) ) > 11 ]
names.tested <-
names.tested[
grep(
x = names.tested,
pattern = "2010"
)
]
names.tested <-
names.tested[
!grepl(
x = names.tested,
pattern = "(change)|(percent)|(resident population)|(male)",
ignore.case = TRUE
)
]
data.test <- data
tmp <-
grepl(
x = names.tested,
pattern = "(population of)|(alone population)",
ignore.case = TRUE
)
names.to.pcnt <- names.tested[ tmp ]
data.test[ , names.to.pcnt ] <-
apply(
X = data[ , names.to.pcnt ],
MAR = 2,
FUN =
function( x ) {
y <-
x /
data[ , "Resident population (April 1 - complete count) 2010" ] *
100
return( y )
}
)
names.mapping <-
data.frame(
Name.original = colnames( data.test ),
Name.made = make.names( colnames( data.test ) ),
stringsAsFactors = FALSE
)
names.mapping$"Name.clean" <- names.mapping$"Name.original"
names.mapping$"Name.clean" <-
stringr::str_replace(
string = names.mapping$"Name.clean",
pattern = "Population of one race \\- ",
replacement = ""
)
names.mapping$"Name.clean" <-
stringr::str_replace(
string = names.mapping$"Name.clean",
pattern = " 2010 \\(complete count\\)",
replacement = ""
)
names.mapping$"Name.clean" <-
stringr::str_replace(
string = names.mapping$"Name.clean",
pattern = "Asian alone population \\- ",
replacement = ""
)
rownames( names.mapping ) <- names.mapping$"Name.made"
colnames( data.test ) <- make.names( colnames( data.test ) )
data.test$"Population.per.square.mile.2010" <-
scale( data.test$"Population.per.square.mile.2010" )
names.tested <- make.names( names.tested )
names.tested <-
names.tested[ names.tested != "Population.per.square.mile.2010" ]
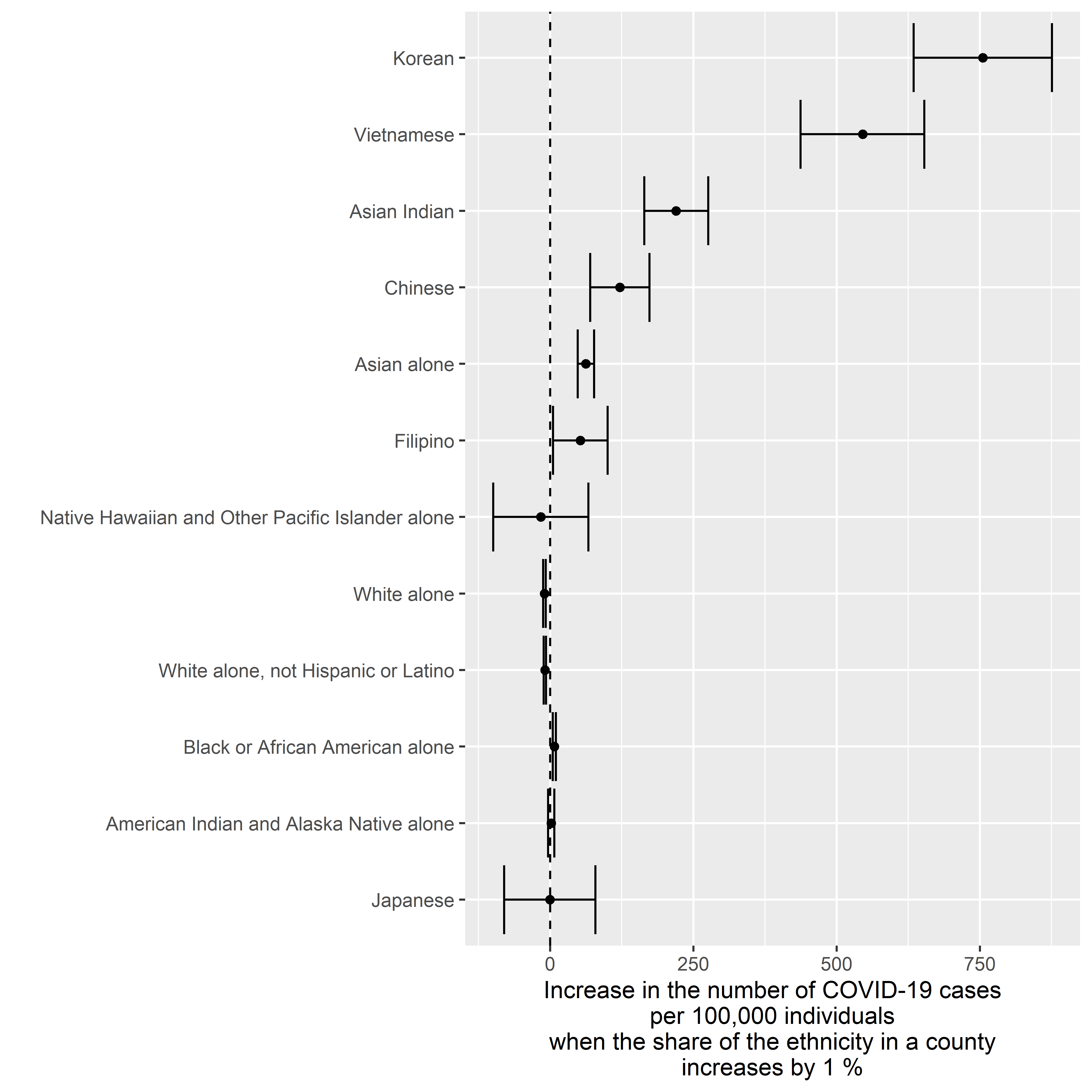
Cases v. Ethnicity
Crude Model
result <-
purrr::map(
.x = names.tested,
.f =
function( x ) {
formula <- paste( "Cases_per_100k ~", x, "+ (1|state)", sep = "" )
formula <- as.formula( formula )
tmp <-
lme4::lmer(
formula = formula,
data = data.test
)
return(
data.frame(
Name = x,
Fixef = lme4::fixef( tmp )[ x ],
lme4::confint.merMod( object = tmp, parm = x )
)
)
}
)
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
result.bound <- dplyr::bind_rows( result, .id = "Variable" )
data.plot <- result.bound
data.plot$"Name.original" <- data.plot$"Name"
data.plot$"Name" <- names.mapping[ data.plot$"Name", "Name.clean" ]
data.plot$"Name" <-
factor(
x = data.plot$"Name",
levels =
data.plot$"Name"[ order( abs( data.plot$"Fixef" ), decreasing = FALSE ) ]
)
ggplot2::ggplot(
data = data.plot,
mapping =
ggplot2::aes(
x = Fixef,
y = Name,
xmin = X2.5..,
xmax = X97.5..
)
) +
ggplot2::geom_point() +
ggplot2::geom_errorbarh() +
ggplot2::geom_vline( xintercept = 0, linetype = "dashed" ) +
ggplot2::ylab( label = "" ) +
ggplot2::xlab( label = "Increase in the number of COVID-19 cases\nper 100,000 individuals\nwhen the share of the ethnicity in a county\nincreases by 1 %" )
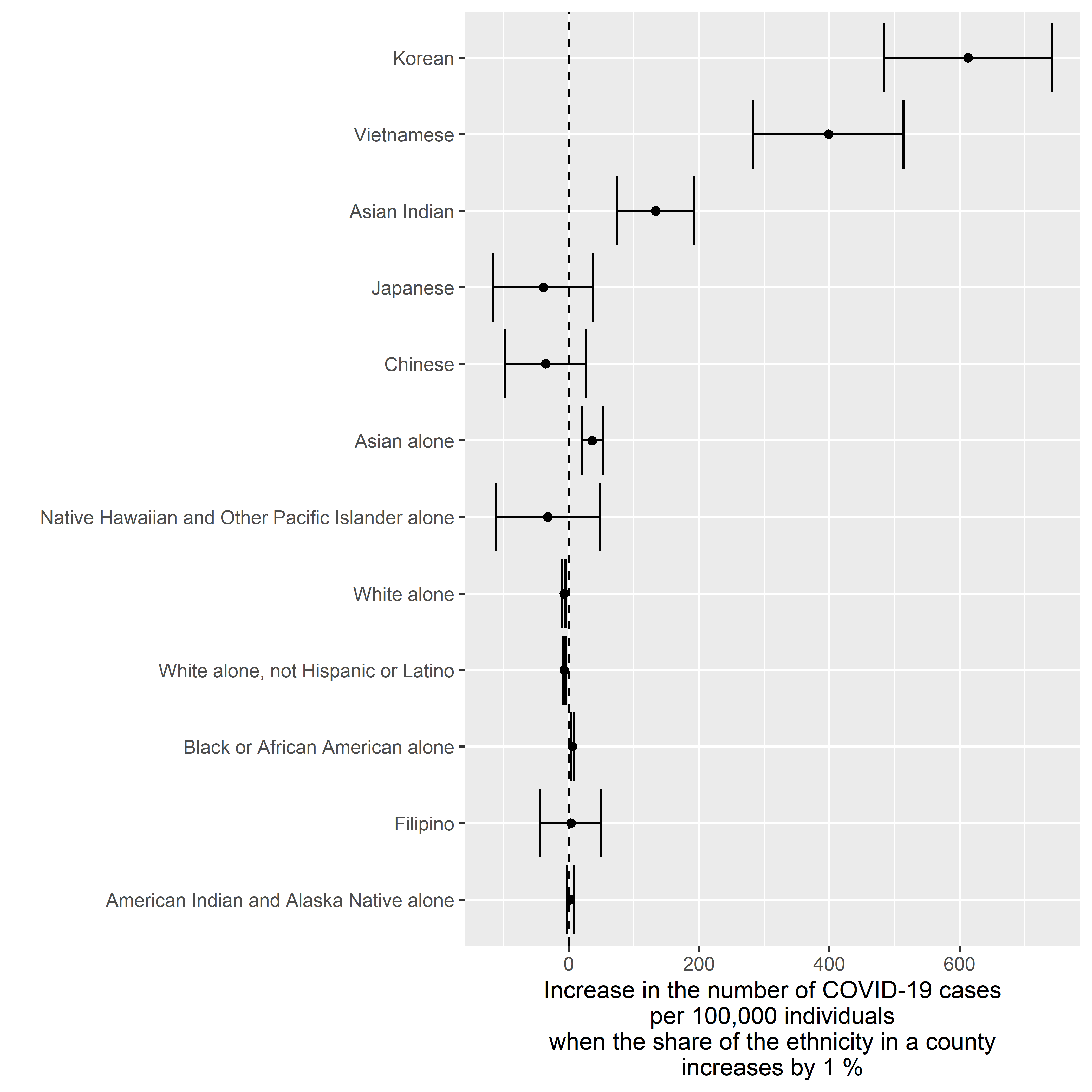
Adjusted for Population Density
result <-
purrr::map(
.x = names.tested,
.f =
function( x ) {
formula <- paste( "Cases_per_100k ~", x, "+ Population.per.square.mile.2010 + (1|state)", sep = "" )
formula <- as.formula( formula )
tmp <-
lme4::lmer(
formula = formula,
data = data.test
)
return(
data.frame(
Name = x,
Fixef = lme4::fixef( tmp )[ x ],
lme4::confint.merMod( object = tmp, parm = x )
)
)
}
)
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
result.bound <- dplyr::bind_rows( result, .id = "Variable" )
data.plot <- result.bound
data.plot$"Name.original" <- data.plot$"Name"
data.plot$"Name" <- names.mapping[ data.plot$"Name", "Name.clean" ]
data.plot$"Name" <-
factor(
x = data.plot$"Name",
levels =
data.plot$"Name"[ order( abs( data.plot$"Fixef" ), decreasing = FALSE ) ]
)
ggplot2::ggplot(
data = data.plot,
mapping =
ggplot2::aes(
x = Fixef,
y = Name,
xmin = X2.5..,
xmax = X97.5..
)
) +
ggplot2::geom_point() +
ggplot2::geom_errorbarh() +
ggplot2::geom_vline( xintercept = 0, linetype = "dashed" ) +
ggplot2::ylab( label = "" ) +
ggplot2::xlab( label = "Increase in the number of COVID-19 cases\nper 100,000 individuals\nwhen the share of the ethnicity in a county\nincreases by 1 %" )
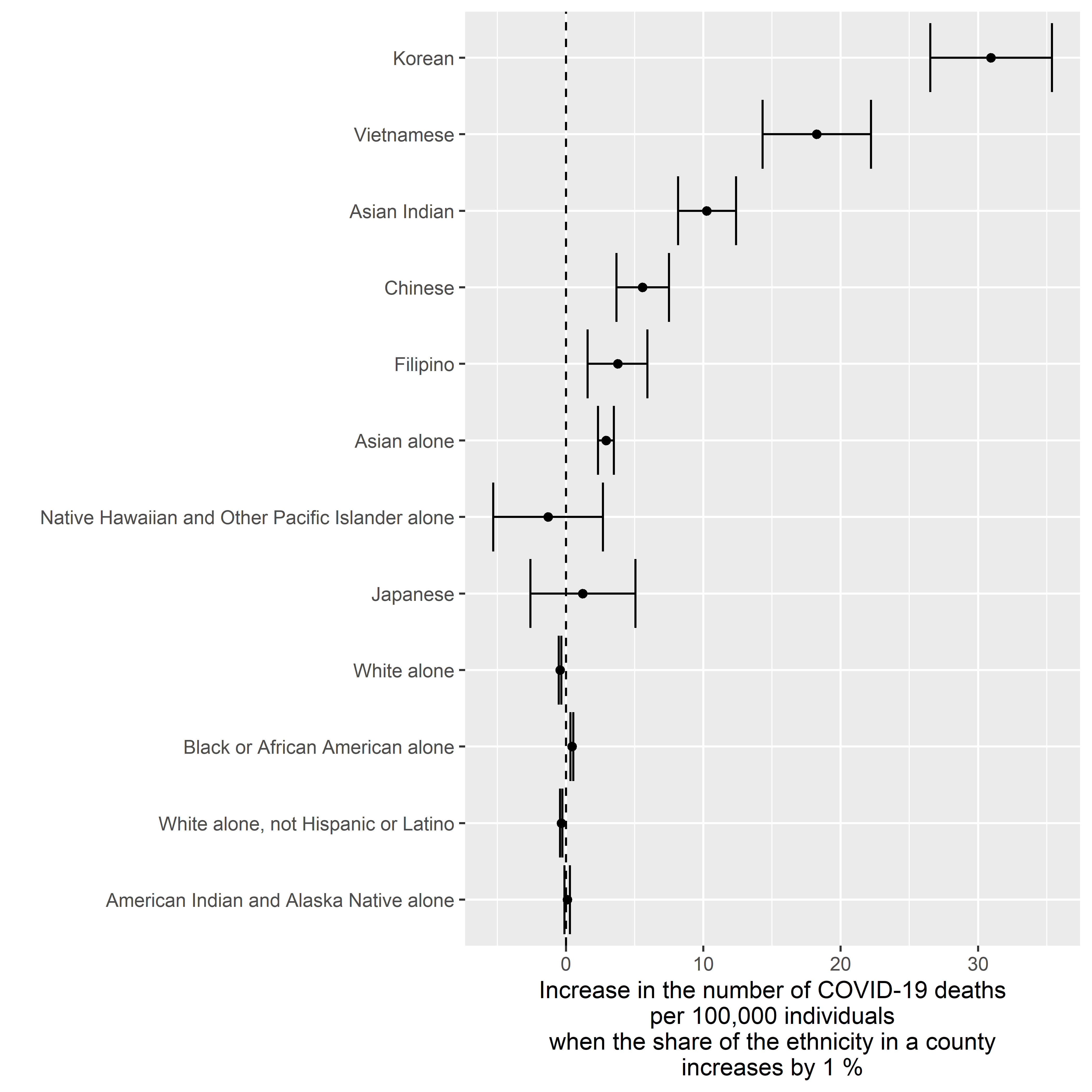
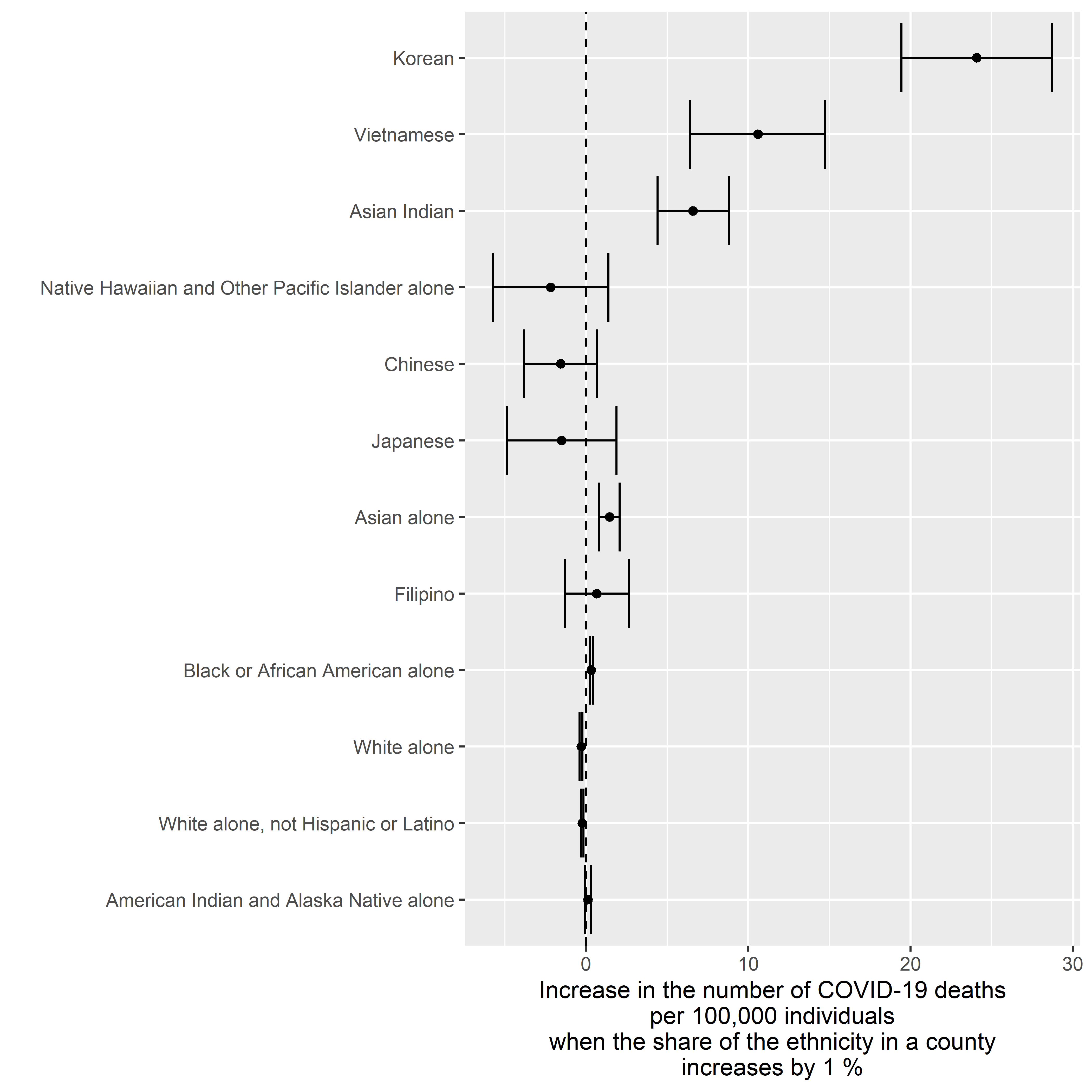
Deaths v. Ethnicity
Crude Model
result <-
purrr::map(
.x = names.tested,
.f =
function( x ) {
formula <- paste( "Deaths_per_100k ~", x, "+ (1|state)", sep = "" )
formula <- as.formula( formula )
tmp <-
lme4::lmer(
formula = formula,
data = data.test
)
return(
data.frame(
Name = x,
Fixef = lme4::fixef( tmp )[ x ],
lme4::confint.merMod( object = tmp, parm = x )
)
)
}
)
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
result.bound <- dplyr::bind_rows( result, .id = "Variable" )
data.plot <- result.bound
data.plot$"Name.original" <- data.plot$"Name"
data.plot$"Name" <- names.mapping[ data.plot$"Name", "Name.clean" ]
data.plot$"Name" <-
factor(
x = data.plot$"Name",
levels =
data.plot$"Name"[ order( abs( data.plot$"Fixef" ), decreasing = FALSE ) ]
)
ggplot2::ggplot(
data = data.plot,
mapping =
ggplot2::aes(
x = Fixef,
y = Name,
xmin = X2.5..,
xmax = X97.5..
)
) +
ggplot2::geom_point() +
ggplot2::geom_errorbarh() +
ggplot2::geom_vline( xintercept = 0, linetype = "dashed" ) +
ggplot2::ylab( label = "" ) +
ggplot2::xlab( label = "Increase in the number of COVID-19 deaths\nper 100,000 individuals\nwhen the share of the ethnicity in a county\nincreases by 1 %" )
Adjusted for Population Density
result <-
purrr::map(
.x = names.tested,
.f =
function( x ) {
formula <- paste( "Deaths_per_100k ~", x, "+ Population.per.square.mile.2010 + (1|state)", sep = "" )
formula <- as.formula( formula )
tmp <-
lme4::lmer(
formula = formula,
data = data.test
)
return(
data.frame(
Name = x,
Fixef = lme4::fixef( tmp )[ x ],
lme4::confint.merMod( object = tmp, parm = x )
)
)
}
)
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
## Computing profile confidence intervals ...
result.bound <- dplyr::bind_rows( result, .id = "Variable" )
data.plot <- result.bound
data.plot$"Name.original" <- data.plot$"Name"
data.plot$"Name" <- names.mapping[ data.plot$"Name", "Name.clean" ]
data.plot$"Name" <-
factor(
x = data.plot$"Name",
levels =
data.plot$"Name"[ order( abs( data.plot$"Fixef" ), decreasing = FALSE ) ]
)
ggplot2::ggplot(
data = data.plot,
mapping =
ggplot2::aes(
x = Fixef,
y = Name,
xmin = X2.5..,
xmax = X97.5..
)
) +
ggplot2::geom_point() +
ggplot2::geom_errorbarh() +
ggplot2::geom_vline( xintercept = 0, linetype = "dashed" ) +
ggplot2::ylab( label = "" ) +
ggplot2::xlab( label = "Increase in the number of COVID-19 deaths\nper 100,000 individuals\nwhen the share of the ethnicity in a county\nincreases by 1 %" )
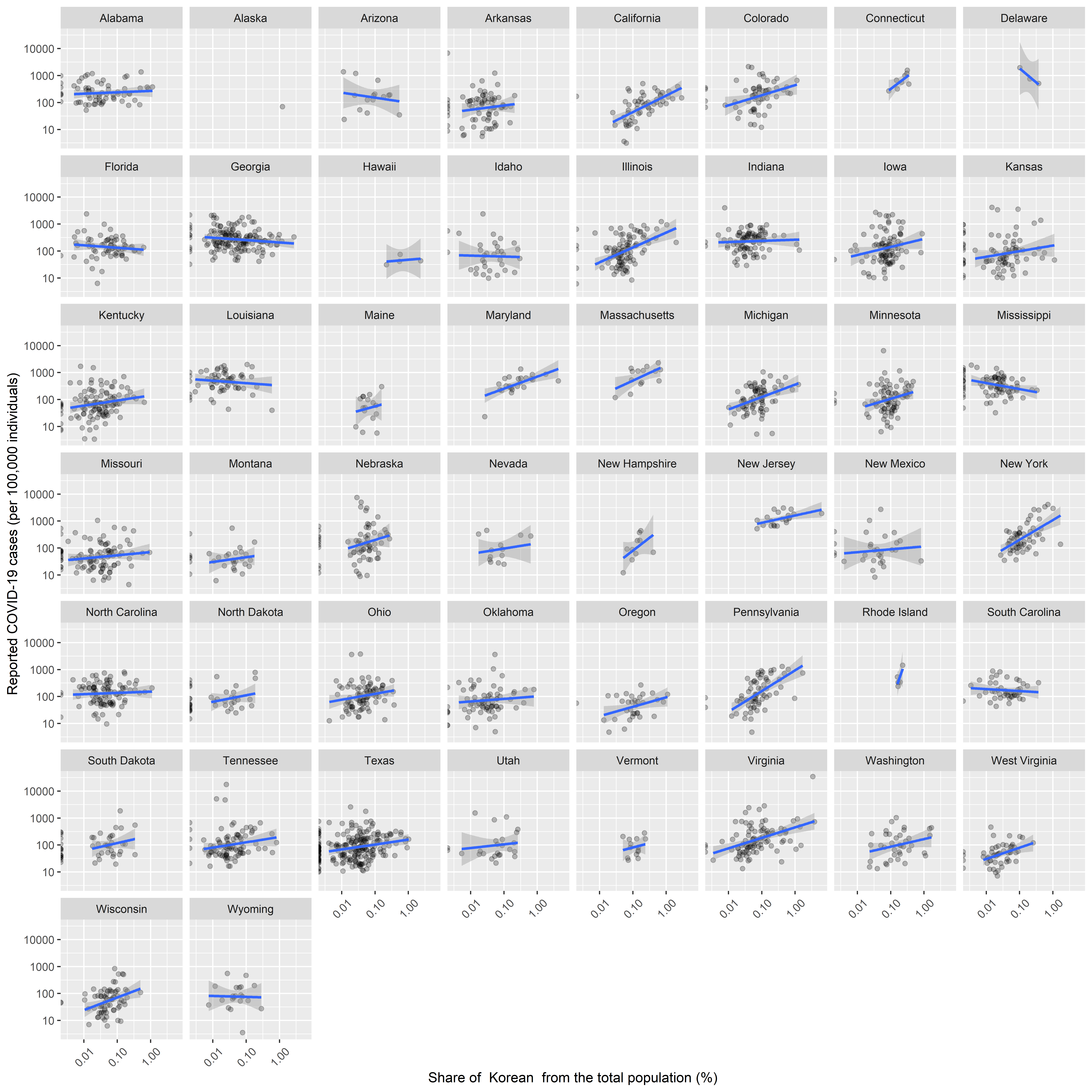
Scatter Plots
Cases vs. Share of Population – By Ethnicity and State
names.figure <- "Asian.alone.population...Korean.2010..complete.count."
for ( i in 1:length( names.figure ) ) {
plot.i <-
ggplot2::ggplot(
data = data.test,
mapping =
ggplot2::aes_string(
x = names.figure[ i ],
y = "Cases_per_100k"
)
) +
ggplot2::geom_point( alpha = 0.25 ) +
ggplot2::geom_smooth( method = "lm" ) +
ggplot2::scale_x_log10() +
ggplot2::scale_y_log10() +
ggplot2::ylab( label = "Reported COVID-19 cases (per 100,000 individuals)" ) +
ggplot2::theme(
axis.text.x =
ggplot2::element_text(
angle = 45,
vjust = 0.5
)
) +
ggplot2::facet_wrap( facets = ggplot2::vars( state ) ) +
ggplot2::xlab(
label =
paste(
"Share of ",
names.mapping[ names.figure[ i ], "Name.clean" ],
" from the total population (%)"
)
)
print( plot.i )
}
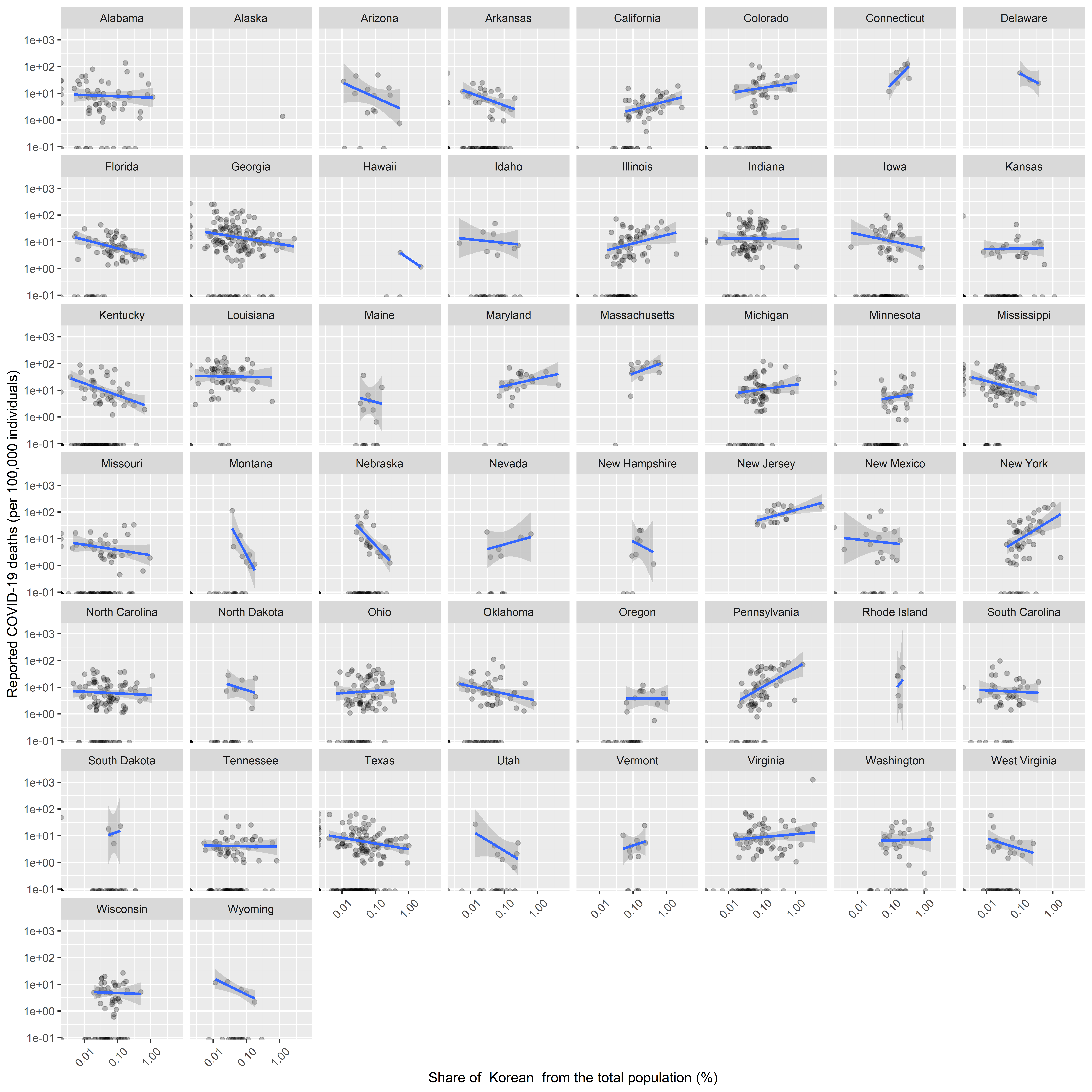
Deaths vs. Share of Population – By Ethnicity and State
names.figure <- "Asian.alone.population...Korean.2010..complete.count."
for ( i in 1:length( names.figure ) ) {
plot.i <-
ggplot2::ggplot(
data = data.test,
mapping =
ggplot2::aes_string(
x = names.figure[ i ],
y = "Deaths_per_100k"
)
) +
ggplot2::geom_point( alpha = 0.25 ) +
ggplot2::geom_smooth( method = "lm" ) +
ggplot2::scale_x_log10() +
ggplot2::scale_y_log10() +
ggplot2::ylab( label = "Reported COVID-19 deaths (per 100,000 individuals)" ) +
ggplot2::theme(
axis.text.x =
ggplot2::element_text(
angle = 45,
vjust = 0.5
)
) +
ggplot2::facet_wrap( facets = ggplot2::vars( state ) ) +
ggplot2::xlab(
label =
paste(
"Share of ",
names.mapping[ names.figure[ i ], "Name.clean" ],
" from the total population (%)"
)
)
print( plot.i )
}